Research
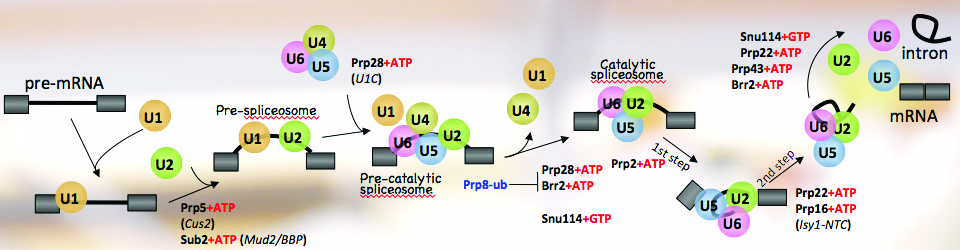
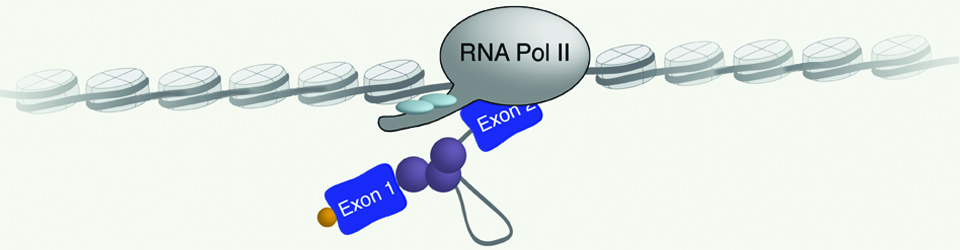
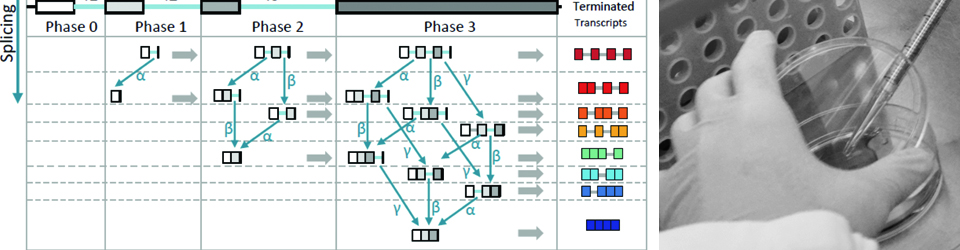
Regulation of gene expression is crucial for every function carried out by the cell, from cell growth and proliferation to the ability of the cell to respond to its ever-changing environment. Hence, understanding cellular function and dysfunction is dependent upon deciphering these gene regulatory mechanisms. This is particularly challenging in the case of eukaryotic genes, which are often interrupted by long stretches of noncoding sequences (introns). These are removed from the newly synthesized RNA, and the remaining sequences (exons) are ligated together to form a mature messenger RNA. This process, pre-messenger RNA splicing, is carried out by the spliceosome made up of 5 small nuclear RNAs and over 100 proteins. The spliceosome undergoes dynamic and coordinated rearrangements in order to recognize splicing signals in the RNA and catalyze the splicing reaction. Remarkably, the spliceosome assembles onto the pre-mRNA co-transcriptionally, while the RNA polymerase is actively engaged with the chromatin template. This close spatial and temporal proximity of splicing and transcription raise the intriguing possibility that assembly of the spliceosome onto pre-mRNA may be influenced by transcription, and/or the state of the chromatin and vice versa; splicing may influence transcription and chromatin modification. The goal of our research is to decipher the workings of this elegant ribonucleoprotein machine. Moreover, we seek to understand how regulation of RNA splicing and other RNA processing reactions allows the cell to respond to its environment.
Funding
NIH/NIGMS, NSF, Whitcome, HHMI, Keith and Cecilia Terasaki Endowed Chair.